This research focuses on targeting transcriptional kinases CDK11, CDK12/13, and autophagy ULK1 kinase, among others, for cancer therapy and is supported by the National Cancer Institute and the Department of Defense's Congressionally Directed Medical Research Programs Breast Cancer Research Program. Our efforts are twofold here: (1) the development of selective, in vivo active probes to study corresponding biology and (2) hit-to-lead and lead optimizations for further preclinical development.
Another critical area of our research involves the targeted protein degradation strategy, where we focus on the rational discovery of molecular glue degraders. This approach facilitates the degradation of specific proteins via the ubiquitin-proteasome system, offering a novel method to target traditionally 'undruggable' proteins within the cancer proteome.
We also spearhead initiatives in chemoproteomics-enabled screening combined with fragment-based drug discovery, funded by multiple R21 grants from NCI and the Live Like Bella Pediatric Cancer Biomedical Research Program (State of Florida). This approach enables the identification and targeting of cancer-specific biomolecular interactions, supporting the advancement of personalized cancer therapies
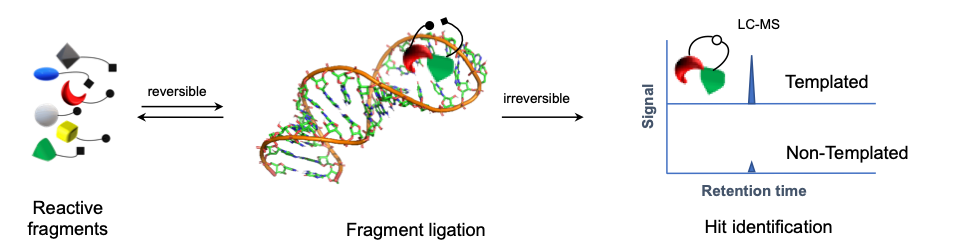
Kinetic Target-Guided Synthesis (TGS) strategy is a powerful yet unconventional approach in which the biological target is actively engaged in the design and the synthesis of its own inhibitory compounds in the irreversible fashion. Fragments bind to the biological target – a protein or DNA/RNA fragment sites – simultaneously with reacting groups positioned within conformational reach of each other, increasing their effective molarity. Although kinetic TGS was conceptually proposed in the 1980s, it is still relatively unexplored in comparison to other fragment-based approaches.